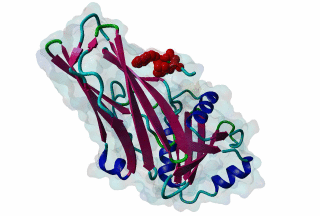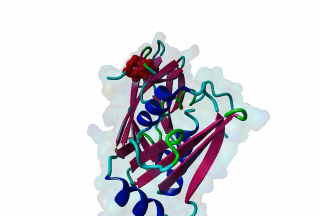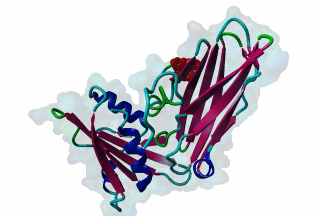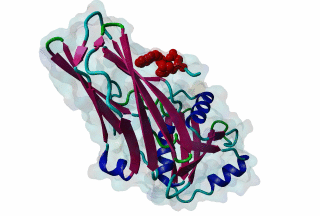
LIG_AP2alpha_2 - PDB:1KY6
iELM
identifying human SLiM-mediated interaction
Background
Short linear motifs (SLiMs) are on average 3-10 amino acids long and bind onto the surface of a globular domain. Each SLiM has 2-5 residues essential for function which are defined by the ELM resource in the form of regular expressions. SLiMs are enriched in regions of intrinsic disorder (IDRs) and often form secondary structure upon binding. SLiMs short nature and locality within IDRs means they can be formed stochastically by convergent evolution.
SLiM are involved in a range of functions, including as sites for post-translational modifications, sites for cleavage, targeting signals for cellular compartments and protein interaction modules. These are all mediated by their interaction interfaces.
The growing knowledge of protein-protein interactions and the importance of SLiMs in protein regulation encouraged us to produce a method to identify these interfaces within networks. We used this method to undertake a survey of the human interactome. To find the results for your protein of interest or ELM class of interest please use the Search tab at the top of the page
For more details on the method please refer to the about section or to the following paper:
RJ Weatheritt, K Luck, E Petsalaki, NE Davey, TJ Gibson. (2011) The identification of short linear motif-mediated interfaces within the human interactome